COMBAT-TB
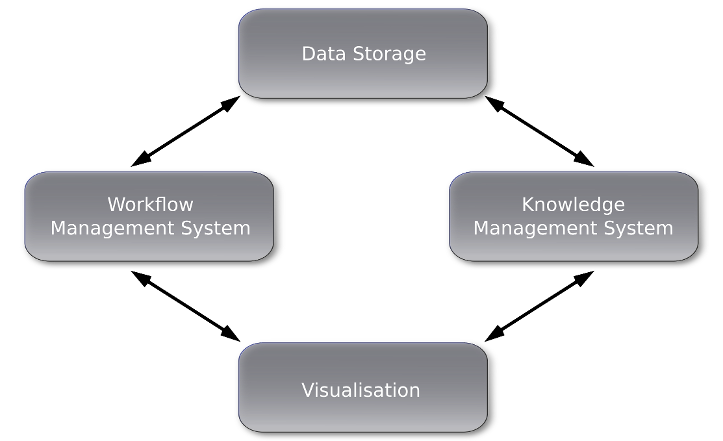
COMBAT-TB is a SAMRC Flagship and DST/NRF Research Chair funded project undertaken by researchers and software developers at SANBI that provides tools for understanding host-pathogen interactions. While this platform is targeted at Tuberculosis research, the goal is to deploy similar computational tools for other pathogens such as Listeriosis. Our focus is on two related software development initiatives:
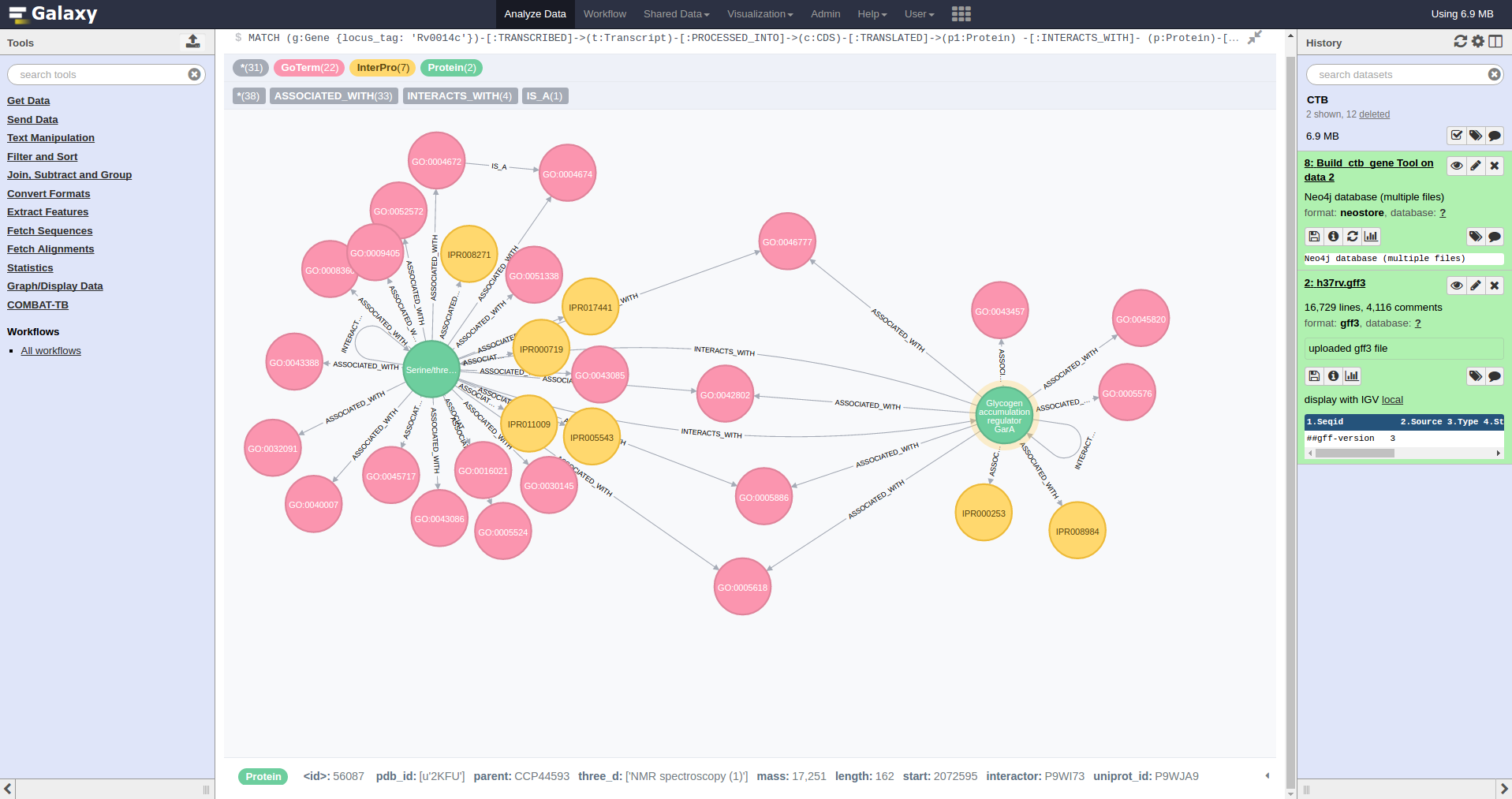
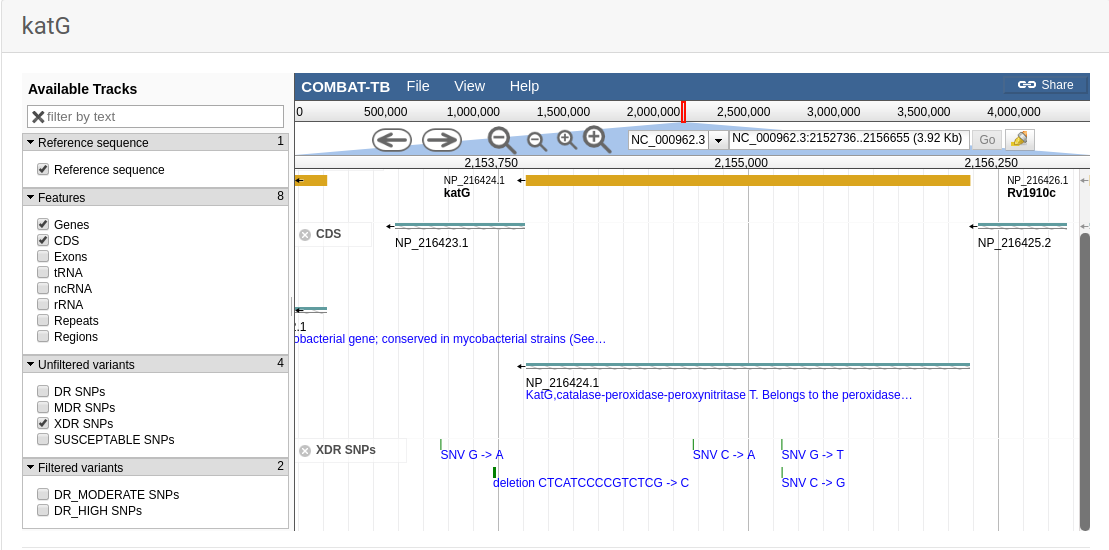
COMBAT-TB eXplorer, a comprehensive knowledgebase of the M. tuberculosis genome and associated information.
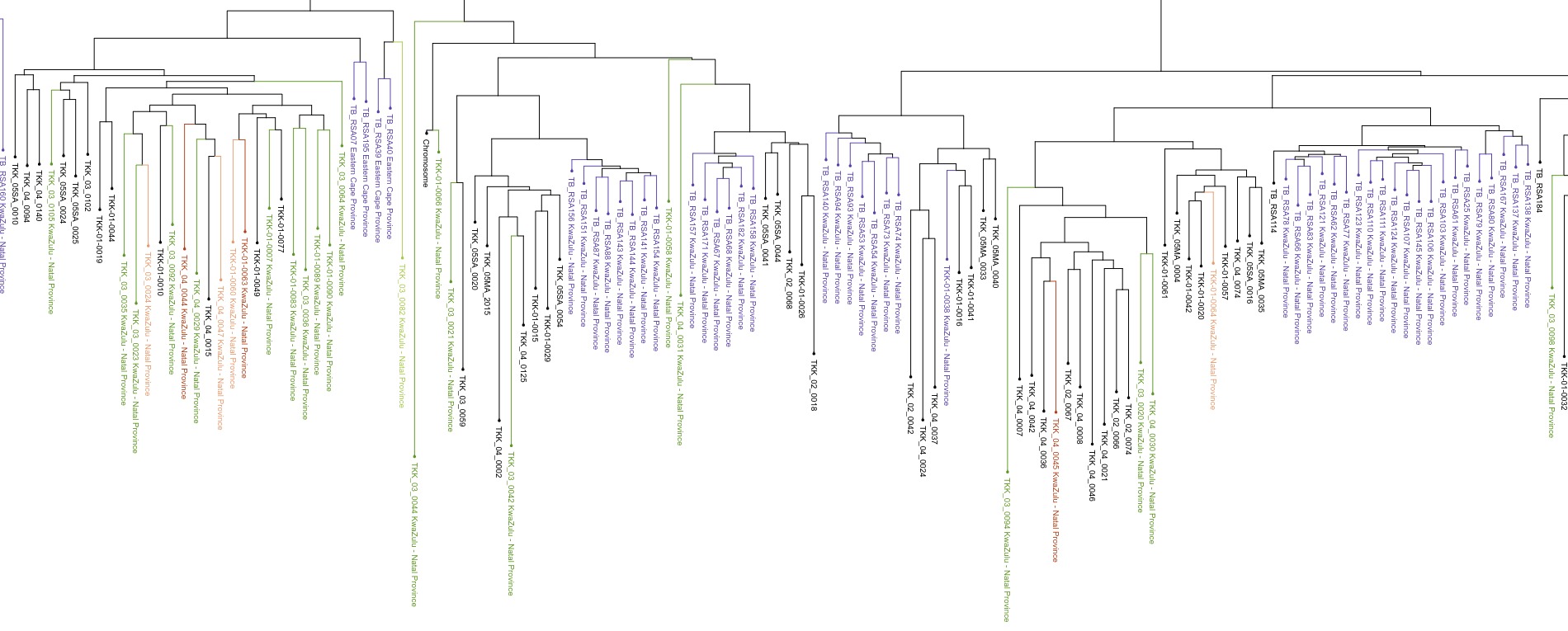
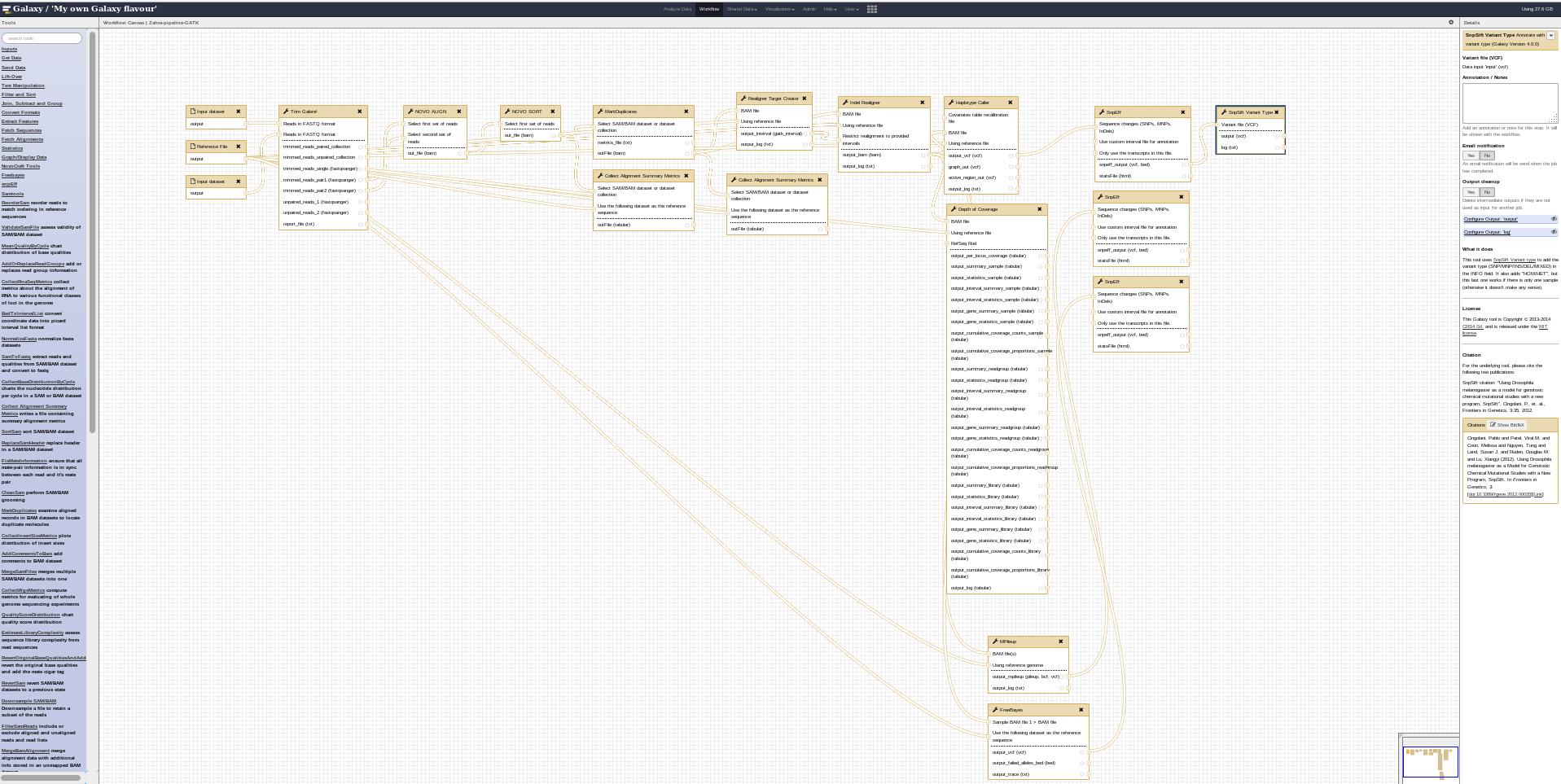
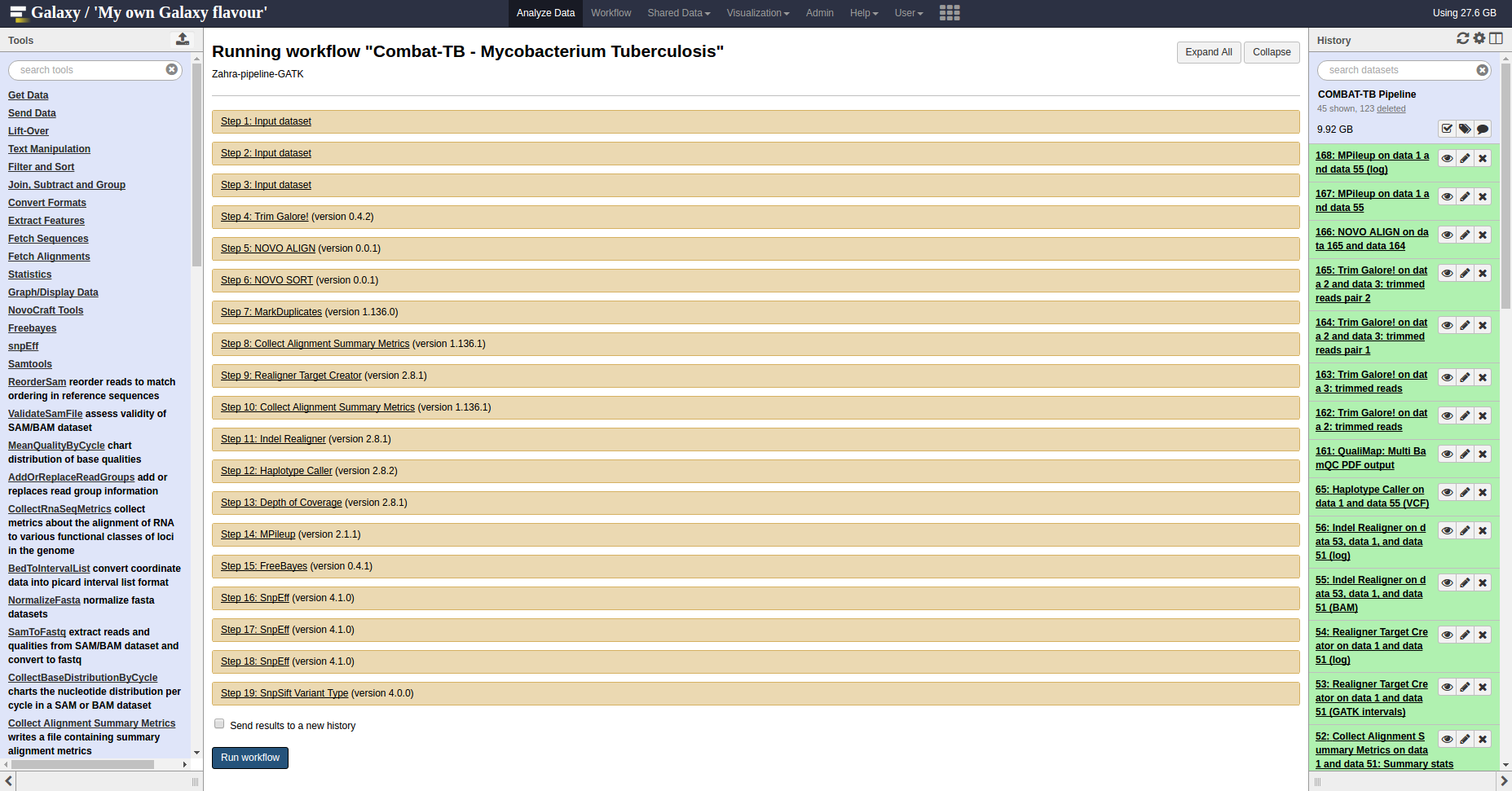
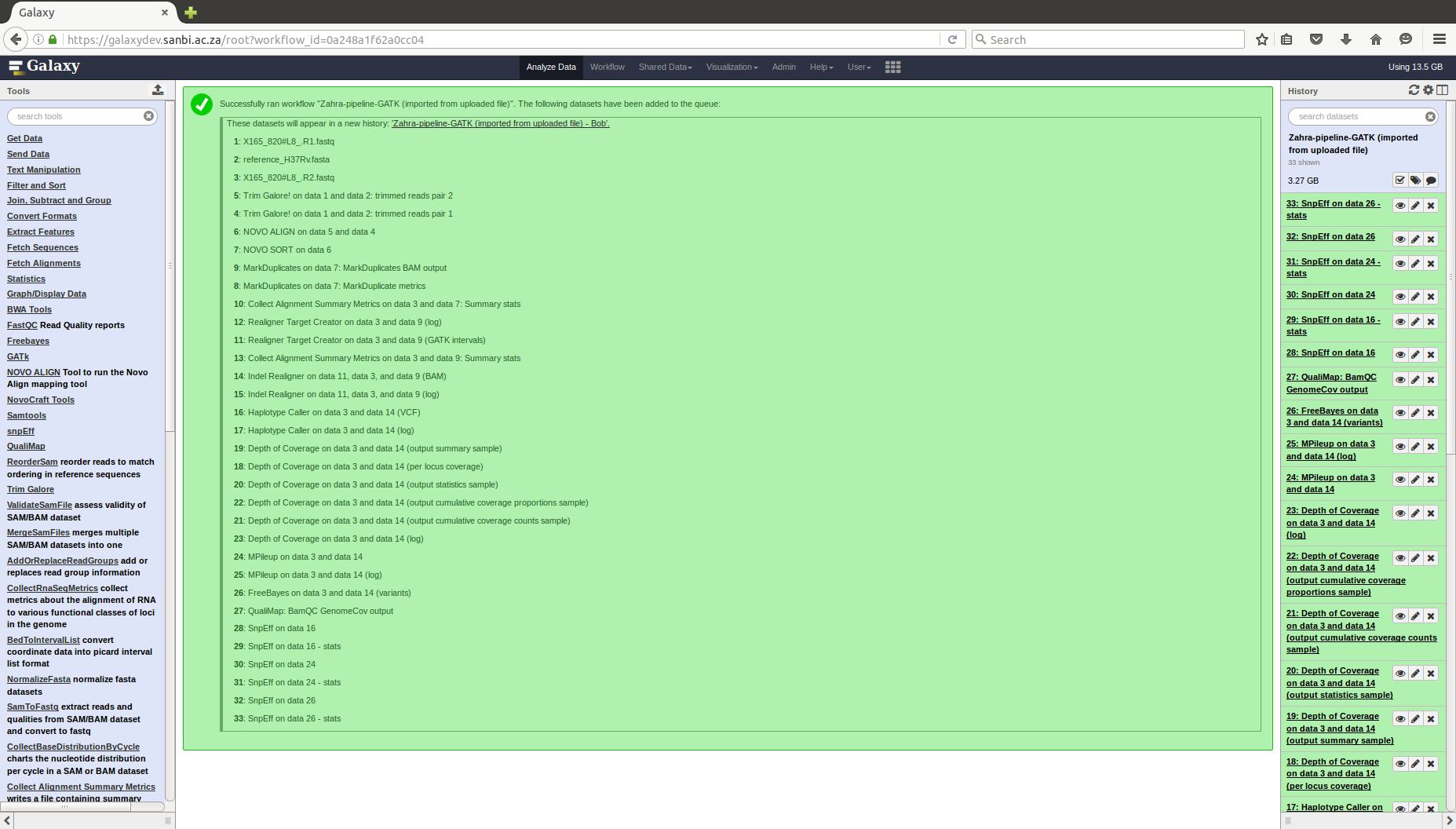
COMBAT-TB Workbench, a set of tools and workflows for M. tuberculosis sequence data analysis.